Context Matters for Foundation Models in Biology
August 16, 2024Just as words can have multiple meanings depending on the context of a sentence, proteins can play different roles in a cell based on their cellular environments. Advances in our understanding of protein and biomolecule functions have been propelled by recent breakthroughs in transformer-based models, such as large language models and generative pre-trained transformers, which automatically learn word semantics from diverse language contexts. Innovating a similar approach for protein functions—viewing them as distributions across various cellular contexts—could enhance the use of foundation models in biology. This would allow the models to dynamically adjust their outputs based on the biological contexts in which they operate. To this end, we have developed PINNACLE, a novel contextual AI model for single-cell biology that supports a broad array of biomedical AI tasks by tailoring its outputs to the cell type context in which the model is asked to make predictions.
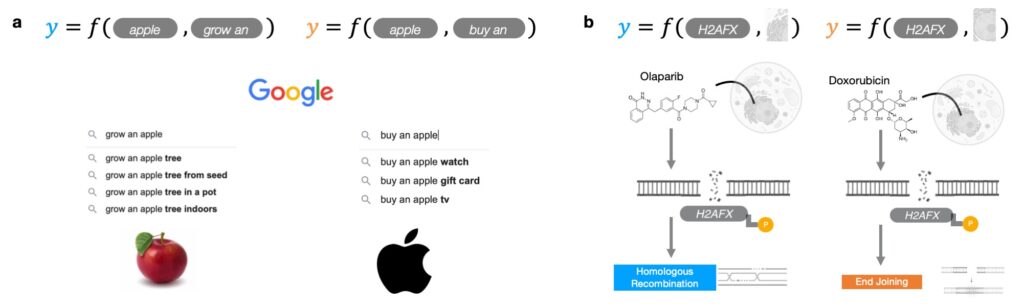
To glean the meaning of a word, we examine nearby words for context clues. For example, “buy an apple” and “grow an apple” yield different recommendations: the first phrase is used to refer to apple products, whereas the second is better associated with apple trees (Figure 1a). To resolve the role of a protein, we interrogate it in the context of the proteins with which it interacts and the cells in which it exists. For instance, H2AFX is a gene that can be involved in homologous recombination or end joining depending on its cellular context (Figure 1b).
Cellular context is critical to understanding protein function and developing molecular therapies. Still, modeling proteins across biological contexts, such as the cell types that they are activated in and the proteins they interact with, remains an algorithmic challenge. Current approaches are context-free: learning on a reference context-agnostic dataset, a single context at a time, or an integrated summary across multiple contexts. As a result, they cannot tailor outputs based on a given context, which can lead to poor predictive performance when applied to a context-specific setting or a never-before-seen context. We develop PINNACLE, a new geometric deep learning approach that generates context-aware protein representations to address these challenges.
Context-specific geometric deep learning PINNACLE model
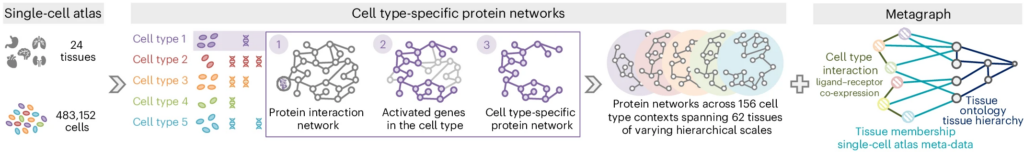
PINNACLE is a novel geometric deep learning model that learns on contextualized protein interaction networks to produce 394,760 protein representations from 156 cell type contexts across 24 tissues. By leveraging a multi-organ single-cell atlas Tabula Sapiens from CZ CELLxGENE Discover, we construct 156 cell type specific protein interaction networks that are maximally similar to the global reference protein interaction network while maintaining cell type specificity (left and middle panels of Figure 2). We additionally create a metagraph to capture the tissue hierarchy and cell type communication among the cell type specific protein interaction networks (right panel of Figure 2). There are four distinct edge types in the metagraph: cell type to cell type (i.e., cell type interaction), cell type to tissue, tissue to cell type (i.e., tissue membership of the cell type), and tissue to tissue (i.e., parent-child tissue relationship in a tissue ontology). This results in multi-scale networks representing protein, cell type, and tissue information in a unified data representation.
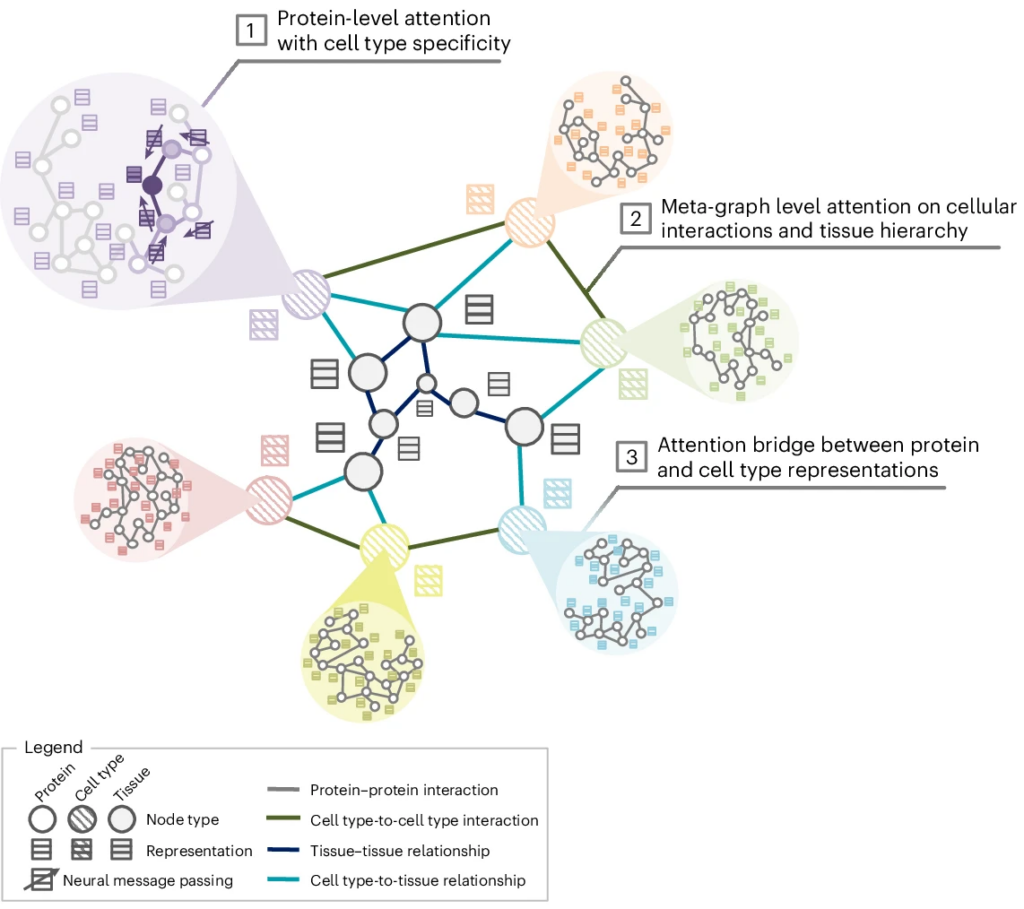
PINNACLE’s algorithm specifies graph neural message passing transformations on multi-scale protein interaction networks. It performs neural message passing with attention for each cell type specific protein interaction network (component 1 in Figure 3) and metagraph (component 2 in Figure 3), and aligns the protein and cell type embeddings using an attention bridge (component 3 in Figure 3). First, PINNACLE learns a trainable weight matrix, node embeddings, and attention weights for each cell type specific protein interaction network. They are optimized based on two protein-level tasks: link prediction (i.e., whether an edge exists between a pair of proteins) and cell type identity (i.e., which cell type a protein is activated in). Secondly, on the metagraph, PINNACLE learns edge type specific trainable weight matrices, node embeddings, and attention weights and aggregates the edge type specific node embeddings via another attention mechanism. These trainable parameters are optimized using edge type specific link prediction (i.e., whether a specific type of edge exists between a pair of nodes). Thirdly, PINNACLE learns attention weights to bridge protein and cell type embeddings. This attention bridge facilitates the propagation of neural messages from cell types and tissues to the cell type specific protein embeddings. It enables PINNACLE to generate a unified embedding space of proteins, cell types, and tissues. Further, the attention bridge enforces cellular and tissue organization of the latent protein space based on tissue hierarchy and cell type communication, enabling contextualization of protein representations.
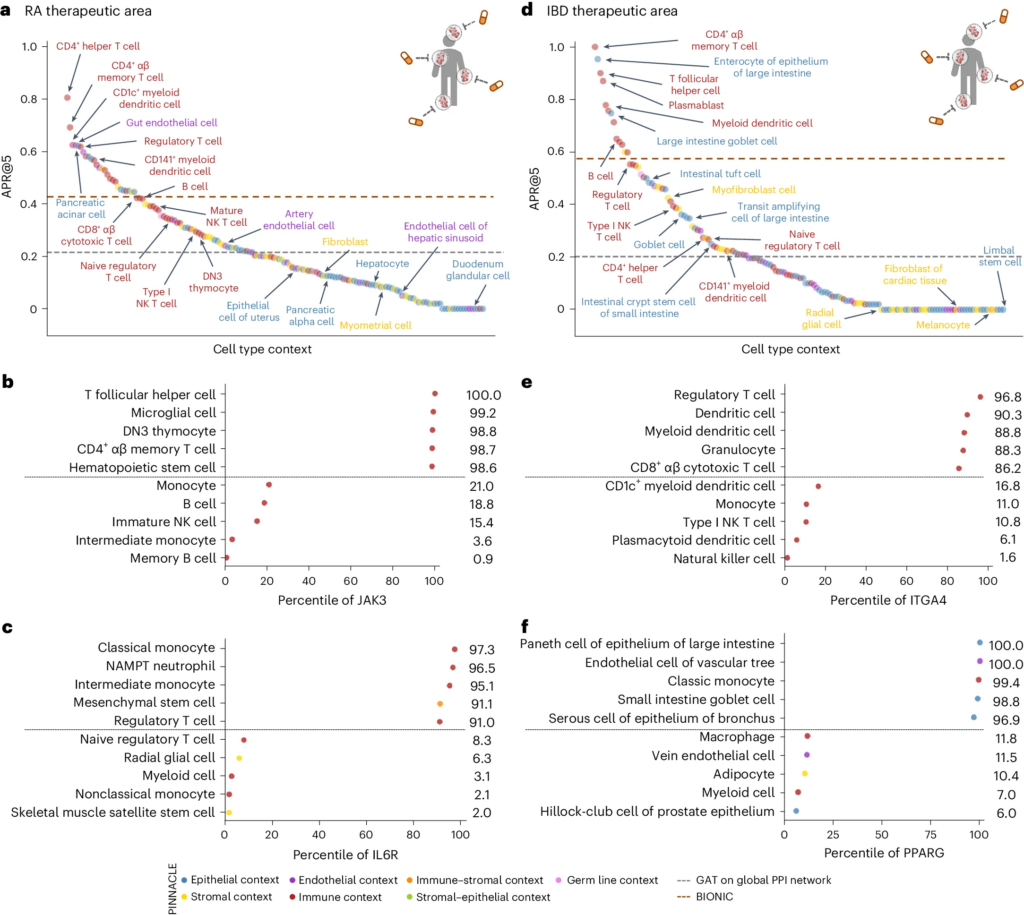
Context-specific predictions
PINNACLE’s contextual representations can be adapted for diverse downstream tasks in which context specificity may play a significant role. Designing safe and effective molecular therapies is one such task that requires understanding the mechanisms of proteins across cell type contexts. We hypothesize that, in contrast to context-free protein representations, contextualized protein representations can enhance 3D structure-based protein representations for resolving immuno-oncological protein interactions (Figure 4) and facilitate the investigation of drugs’ effects across cell types (Figure 5).
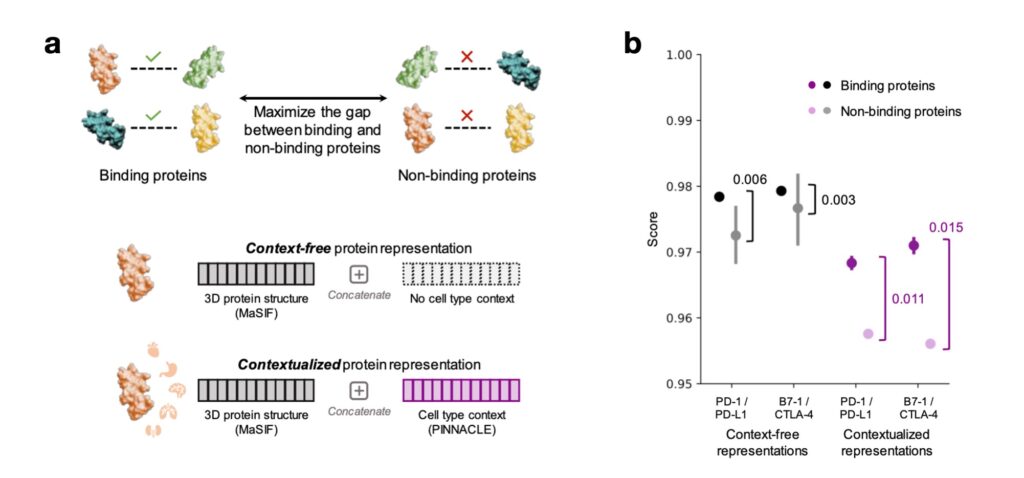
Contextualizing 3D molecular structures of proteins using existing structure-based models is limited by the scarcity of structures captured in context-specific conformations. We show via demonstrative case studies that PINNACLE’s contextualized protein representations can improve structure-based predictions of binding (and non-binding) proteins (Figure 4a). For two immuno-oncological protein interactors, PD-1/PD-L1 and CTLA-4/B7-1, we generate embeddings of each protein using a state-of-the-art structure-based model, MaSIF. We aggregate these structure-based embeddings with a corresponding genomic-based context-free or contextualized protein embedding (i.e., from PINNACLE). By calculating a binding score (i.e., cosine similarity between a pair of protein embeddings), we find that contextualized embeddings enable better differentiation between binding and non-binding proteins (Figure 4b). This zero-shot analysis of contextualizing 3D structure-based representations exemplifies the potential of contextual learning to improve the modeling of molecular structures across biological contexts.
Nominating therapeutic targets with cell type resolution holds the promise of maximizing the efficacy and safety of a candidate drug. However, it is not possible with current models to systematically predict cell type specific therapeutic potential across all proteins and cell type contexts. By finetuning PINNACLE’s contextualized protein representations, we demonstrate that PINNACLE outperforms state-of-the-art, yet context-free, models in nominating therapeutic targets for rheumatoid arthritis (RA) and inflammatory bowel diseases (IBD). We can pinpoint cell type contexts with higher predictive capability than context-free models (Figure 5a). In collaboration with RA and IBD clinical experts, we find that the most predictive cell types are indeed relevant to RA and IBD. Further, examining predictions of individual proteins across cell types allows us to interrogate each candidate target’s therapeutic potential in each cell type context.
Outlook
PINNACLE is a contextual AI model for representing proteins with cell type resolution. While we demonstrate PINNACLE’s capabilities through cell type specific protein interaction networks, the model can easily be re-trained on any protein network. Our study focuses on single-cell transcriptomic data of healthy individuals, but we expect that training disease-specific PINNACLE models can enable even more accurate predictions of candidate therapeutic targets across cell type contexts. PINNACLE’s ability to adjust its outputs based on the context in which it operates paves the way for large-scale context-specific predictions in biology.
PINNACLE exemplifies the potential of contextual AI to mimic distinctly human behavior, operating within specific contexts. As humans, we naturally consider and utilize context without conscious effort in our daily interactions and decision-making processes. For instance, we adjust our language, tone, and actions based on the environment and the people we interact with. This inherent ability to dynamically adapt to varying contexts is a cornerstone of human intelligence. In contrast, many current biomedical AI models often lack this contextual adaptability. They tend to operate in a static manner, applying the same logic and patterns regardless of differing biological environments. This limitation can hinder their effectiveness and accuracy in biological systems where context plays a crucial role.
By integrating contextual awareness, models such as PINNACLE can transform biomedical AI. We envision that context-aware models will dynamically adjust their outputs based on the specific cellular environments they encounter, leading to more accurate and relevant predictions and insights. This advancement enhances the functionality of AI models in biology and brings them a step closer to emulating the nuanced and adaptable nature of human thought processes.



